Show the code
set.seed(1337)
library("tidymodels")
tidymodels::tidymodels_prefer()
library("tidyposterior")
library("rstanarm")Set seed and load packages.
set.seed(1337)
library("tidymodels")
tidymodels::tidymodels_prefer()
library("tidyposterior")
library("rstanarm")Load data.
data("iris")
iris <- iris |>
tibble::as_tibble() |>
filter(Species != "setosa") |>
droplevels()
iris_split <- initial_split(iris,
strata = Species,
prop = 0.90)
iris_train <- training(iris_split)
iris_test <- testing(iris_split)The reasonings for, and theoretical aspects of, cross-validation (CV) are expected to be already known. In the tidymodels universe, CV is setup as:
iris_folds <- vfold_cv(iris_train,
v = 10)
iris_folds# 10-fold cross-validation
# A tibble: 10 × 2
splits id
<list> <chr>
1 <split [81/9]> Fold01
2 <split [81/9]> Fold02
3 <split [81/9]> Fold03
4 <split [81/9]> Fold04
5 <split [81/9]> Fold05
6 <split [81/9]> Fold06
7 <split [81/9]> Fold07
8 <split [81/9]> Fold08
9 <split [81/9]> Fold09
10 <split [81/9]> Fold10When it comes to modelling, creating and comparing multiple models is essential. This is easily achieved through the use of recipe() from the recipes package. A recipe is a collection of both a fomula and preprocessing steps. Here preprocessing steps is not applied, but could be: log-transforming data, creating dummy variables (e.g. one hot encoding), sum up low occurring categories to handle class imbalance and dimensionality reduction (see ?sec-preprocessing). A series of recipes are creating and combined:
SL_rec <- recipe(Species ~ Sepal.Length,
data = iris_train)
SLW_rec <- recipe(Species ~ Sepal.Length + Sepal.Width,
data = iris_train)
SLW_int_rec <- recipe(Species ~ Sepal.Length + Sepal.Width,
data = iris_train) |>
step_interact(~ Sepal.Length:Sepal.Width)
PL_rec <- recipe(Species ~ Petal.Length,
data = iris_train)
PLW_rec <- recipe(Species ~ Petal.Length + Petal.Width,
data = iris_train)
PLW_int_rec <- recipe(Species ~ Petal.Length + Petal.Width,
data = iris_train) |>
step_interact(~ Petal.Length:Petal.Width)
recipe_list <- list(SL = SL_rec,
SLW = SLW_rec,
SLW_int = SLW_int_rec,
PL = PL_rec,
PLW = PLW_rec,
PLW_int = PLW_int_rec)
lg_models <- workflow_set(preproc = recipe_list,
models = list(logistic = logistic_reg()),
cross = FALSE)Each of the models are fitted with a purr-like workflow function:
# To save the predicted values and used workflows
keep_pred <- control_resamples(save_pred = TRUE,
save_workflow = TRUE)
lg_models <- lg_models |>
workflow_map("fit_resamples",
resamples = iris_folds,
control = keep_pred,
seed = 1337)
lg_models# A workflow set/tibble: 6 × 4
wflow_id info option result
<chr> <list> <list> <list>
1 SL_logistic <tibble [1 × 4]> <opts[2]> <rsmp[+]>
2 SLW_logistic <tibble [1 × 4]> <opts[2]> <rsmp[+]>
3 SLW_int_logistic <tibble [1 × 4]> <opts[2]> <rsmp[+]>
4 PL_logistic <tibble [1 × 4]> <opts[2]> <rsmp[+]>
5 PLW_logistic <tibble [1 × 4]> <opts[2]> <rsmp[+]>
6 PLW_int_logistic <tibble [1 × 4]> <opts[2]> <rsmp[+]>Two simple evaluation metrics for logistic regressions are the accuracy and Area Under the Curve (AUC). It is implicit that it is area under the Receiver Operating Characteristic (ROC) curve. To view these two evaluation metrics (the column mean):
collect_metrics(lg_models) |>
select(-c(.config, preproc, .estimator)) |>
filter(.metric == "accuracy") |>
arrange(desc(mean))# A tibble: 6 × 6
wflow_id model .metric mean n std_err
<chr> <chr> <chr> <dbl> <int> <dbl>
1 PL_logistic logistic_reg accuracy 0.933 10 0.0296
2 PLW_logistic logistic_reg accuracy 0.933 10 0.0246
3 PLW_int_logistic logistic_reg accuracy 0.933 10 0.0246
4 SL_logistic logistic_reg accuracy 0.756 10 0.0363
5 SLW_logistic logistic_reg accuracy 0.711 10 0.0444
6 SLW_int_logistic logistic_reg accuracy 0.711 10 0.0474collect_metrics(lg_models) |>
select(-c(.config, preproc, .estimator)) |>
filter(.metric == "roc_auc") |>
arrange(desc(mean))# A tibble: 6 × 6
wflow_id model .metric mean n std_err
<chr> <chr> <chr> <dbl> <int> <dbl>
1 PLW_logistic logistic_reg roc_auc 0.989 10 0.00705
2 PLW_int_logistic logistic_reg roc_auc 0.989 10 0.00705
3 PL_logistic logistic_reg roc_auc 0.987 10 0.00923
4 SL_logistic logistic_reg roc_auc 0.804 10 0.0633
5 SLW_logistic logistic_reg roc_auc 0.798 10 0.0664
6 SLW_int_logistic logistic_reg roc_auc 0.795 10 0.0737 To illustrate the two metrics:
lg_models |>
autoplot(metric = "accuracy") +
geom_label(aes(label = wflow_id)) +
theme(legend.position = "none") +
xlim(c(0.5, 6.5)) +
ggtitle("Accuracy Stratified on Model") +
theme(text=element_text(size=13))
lg_models |>
collect_predictions() |>
group_by(wflow_id) |>
roc_curve(truth = Species,
.pred_versicolor) |>
autoplot() +
ggtitle("ROC Rurve Stratified on Model") +
theme(text=element_text(size=12))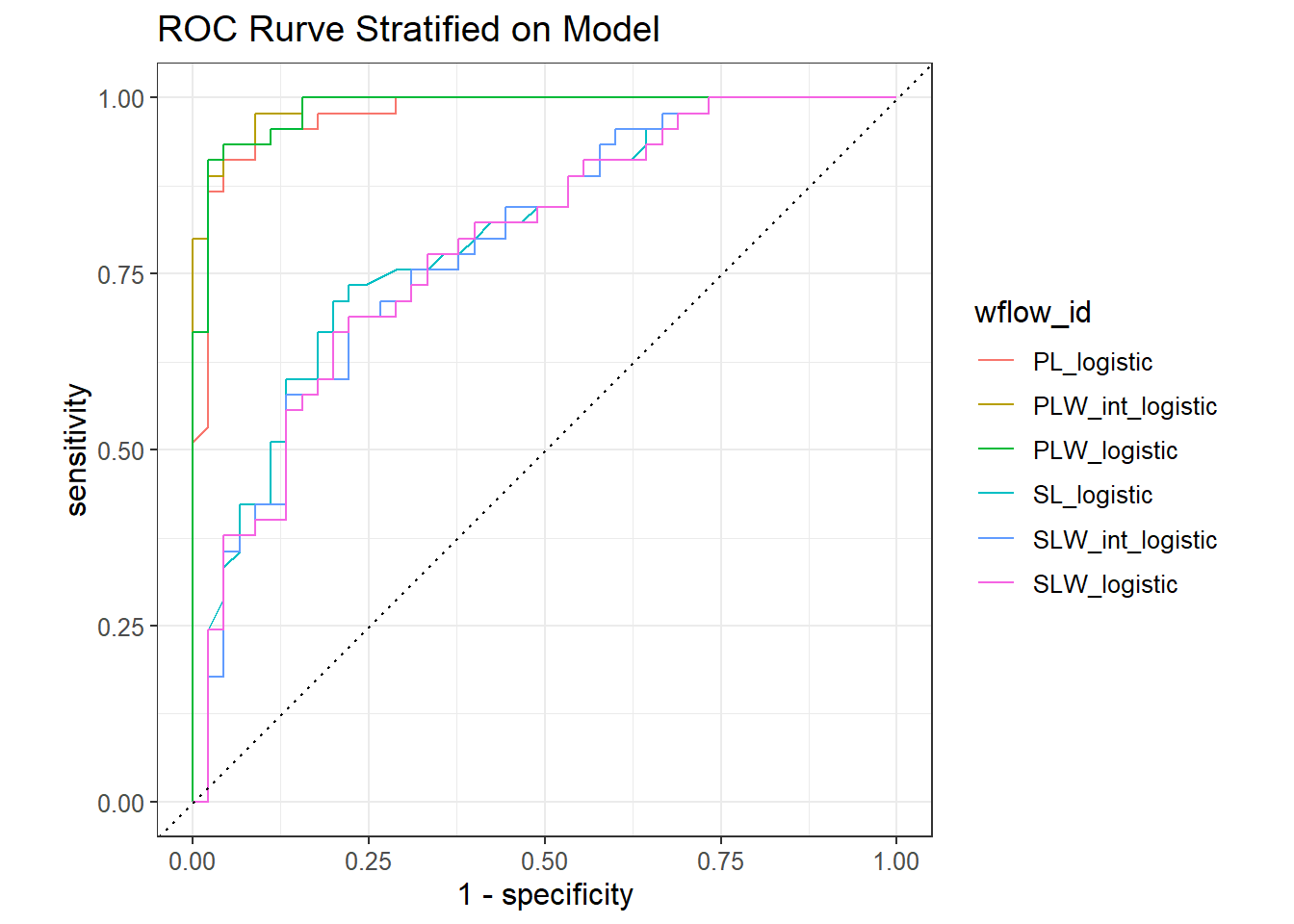
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.3 (2024-02-29 ucrt)
os Windows 11 x64 (build 22631)
system x86_64, mingw32
ui RTerm
language (EN)
collate English_United Kingdom.utf8
ctype English_United Kingdom.utf8
tz Europe/Copenhagen
date 2024-05-30
pandoc 3.1.11 @ C:/Program Files/RStudio/resources/app/bin/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
! package * version date (UTC) lib source
abind 1.4-5 2016-07-21 [1] CRAN (R 4.3.1)
backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.1)
base64enc 0.1-3 2015-07-28 [1] CRAN (R 4.3.1)
bayesplot 1.11.1 2024-02-15 [1] CRAN (R 4.3.3)
boot 1.3-29 2024-02-19 [2] CRAN (R 4.3.3)
broom * 1.0.5 2023-06-09 [1] CRAN (R 4.3.3)
cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.3)
checkmate 2.3.1 2023-12-04 [1] CRAN (R 4.3.3)
class 7.3-22 2023-05-03 [2] CRAN (R 4.3.3)
cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.3)
codetools 0.2-19 2023-02-01 [2] CRAN (R 4.3.3)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.3)
colourpicker 1.3.0 2023-08-21 [1] CRAN (R 4.3.3)
conflicted 1.2.0 2023-02-01 [1] CRAN (R 4.3.3)
crosstalk 1.2.1 2023-11-23 [1] CRAN (R 4.3.3)
data.table 1.15.4 2024-03-30 [1] CRAN (R 4.3.3)
dials * 1.2.1 2024-02-22 [1] CRAN (R 4.3.3)
DiceDesign 1.10 2023-12-07 [1] CRAN (R 4.3.3)
digest 0.6.35 2024-03-11 [1] CRAN (R 4.3.3)
distributional 0.4.0 2024-02-07 [1] CRAN (R 4.3.3)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.2)
DT 0.33 2024-04-04 [1] CRAN (R 4.3.3)
dygraphs 1.1.1.6 2018-07-11 [1] CRAN (R 4.3.3)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.3)
evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.3)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.3)
farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.3)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.3)
foreach 1.5.2 2022-02-02 [1] CRAN (R 4.3.3)
furrr 0.3.1 2022-08-15 [1] CRAN (R 4.3.3)
future 1.33.2 2024-03-26 [1] CRAN (R 4.3.3)
future.apply 1.11.2 2024-03-28 [1] CRAN (R 4.3.3)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.3)
ggplot2 * 3.5.1 2024-04-23 [1] CRAN (R 4.3.3)
globals 0.16.3 2024-03-08 [1] CRAN (R 4.3.3)
glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.3)
gower 1.0.1 2022-12-22 [1] CRAN (R 4.3.1)
GPfit 1.0-8 2019-02-08 [1] CRAN (R 4.3.3)
gridExtra 2.3 2017-09-09 [1] CRAN (R 4.3.3)
gtable 0.3.5 2024-04-22 [1] CRAN (R 4.3.3)
gtools 3.9.5 2023-11-20 [1] CRAN (R 4.3.3)
hardhat 1.3.1 2024-02-02 [1] CRAN (R 4.3.3)
htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.3.3)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.3)
httpuv 1.6.15 2024-03-26 [1] CRAN (R 4.3.3)
igraph 2.0.3 2024-03-13 [1] CRAN (R 4.3.3)
infer * 1.0.7 2024-03-25 [1] CRAN (R 4.3.3)
inline 0.3.19 2021-05-31 [1] CRAN (R 4.3.3)
ipred 0.9-14 2023-03-09 [1] CRAN (R 4.3.3)
iterators 1.0.14 2022-02-05 [1] CRAN (R 4.3.3)
jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.3)
knitr 1.46 2024-04-06 [1] CRAN (R 4.3.3)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.1)
later 1.3.2 2023-12-06 [1] CRAN (R 4.3.3)
lattice 0.22-5 2023-10-24 [2] CRAN (R 4.3.3)
lava 1.8.0 2024-03-05 [1] CRAN (R 4.3.3)
lhs 1.1.6 2022-12-17 [1] CRAN (R 4.3.3)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.3)
listenv 0.9.1 2024-01-29 [1] CRAN (R 4.3.3)
lme4 1.1-35.3 2024-04-16 [1] CRAN (R 4.3.3)
loo 2.7.0 2024-02-24 [1] CRAN (R 4.3.3)
lubridate 1.9.3 2023-09-27 [1] CRAN (R 4.3.3)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.3)
markdown 1.12 2023-12-06 [1] CRAN (R 4.3.3)
MASS 7.3-60.0.1 2024-01-13 [2] CRAN (R 4.3.3)
Matrix 1.6-5 2024-01-11 [2] CRAN (R 4.3.3)
matrixStats 1.3.0 2024-04-11 [1] CRAN (R 4.3.3)
memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.3)
mime 0.12 2021-09-28 [1] CRAN (R 4.3.1)
miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.3.3)
minqa 1.2.6 2023-09-11 [1] CRAN (R 4.3.3)
modeldata * 1.3.0 2024-01-21 [1] CRAN (R 4.3.3)
munsell 0.5.1 2024-04-01 [1] CRAN (R 4.3.3)
nlme 3.1-164 2023-11-27 [2] CRAN (R 4.3.3)
nloptr 2.0.3 2022-05-26 [1] CRAN (R 4.3.3)
nnet 7.3-19 2023-05-03 [2] CRAN (R 4.3.3)
parallelly 1.37.1 2024-02-29 [1] CRAN (R 4.3.3)
parsnip * 1.2.1 2024-03-22 [1] CRAN (R 4.3.3)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.3)
pkgbuild 1.4.4 2024-03-17 [1] CRAN (R 4.3.3)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.3)
plyr 1.8.9 2023-10-02 [1] CRAN (R 4.3.3)
posterior 1.5.0 2023-10-31 [1] CRAN (R 4.3.3)
prodlim 2023.08.28 2023-08-28 [1] CRAN (R 4.3.3)
promises 1.3.0 2024-04-05 [1] CRAN (R 4.3.3)
purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.3)
QuickJSR 1.1.3 2024-01-31 [1] CRAN (R 4.3.3)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.3)
Rcpp * 1.0.12 2024-01-09 [1] CRAN (R 4.3.3)
D RcppParallel 5.1.7 2023-02-27 [1] CRAN (R 4.3.3)
recipes * 1.0.10 2024-02-18 [1] CRAN (R 4.3.3)
reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.3.3)
rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.3)
rmarkdown 2.26 2024-03-05 [1] CRAN (R 4.3.3)
rpart 4.1.23 2023-12-05 [2] CRAN (R 4.3.3)
rsample * 1.2.1 2024-03-25 [1] CRAN (R 4.3.3)
rstan 2.32.6 2024-03-05 [1] CRAN (R 4.3.3)
rstanarm * 2.32.1 2024-01-18 [1] CRAN (R 4.3.3)
rstantools 2.4.0 2024-01-31 [1] CRAN (R 4.3.3)
rstudioapi 0.16.0 2024-03-24 [1] CRAN (R 4.3.3)
scales * 1.3.0 2023-11-28 [1] CRAN (R 4.3.3)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.3)
shiny 1.8.1.1 2024-04-02 [1] CRAN (R 4.3.3)
shinyjs 2.1.0 2021-12-23 [1] CRAN (R 4.3.3)
shinystan 2.6.0 2022-03-03 [1] CRAN (R 4.3.3)
shinythemes 1.2.0 2021-01-25 [1] CRAN (R 4.3.3)
StanHeaders 2.32.6 2024-03-01 [1] CRAN (R 4.3.3)
stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.2)
stringr 1.5.1 2023-11-14 [1] CRAN (R 4.3.3)
survival 3.5-8 2024-02-14 [2] CRAN (R 4.3.3)
tensorA 0.36.2.1 2023-12-13 [1] CRAN (R 4.3.2)
threejs 0.3.3 2020-01-21 [1] CRAN (R 4.3.3)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.3)
tidymodels * 1.2.0 2024-03-25 [1] CRAN (R 4.3.3)
tidyposterior * 1.0.1 2023-10-11 [1] CRAN (R 4.3.3)
tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.3)
tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.3.3)
timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.3)
timeDate 4032.109 2023-12-14 [1] CRAN (R 4.3.2)
tune * 1.2.1 2024-04-18 [1] CRAN (R 4.3.3)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.3)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.3)
withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.3)
workflows * 1.1.4 2024-02-19 [1] CRAN (R 4.3.3)
workflowsets * 1.1.0 2024-03-21 [1] CRAN (R 4.3.3)
xfun 0.43 2024-03-25 [1] CRAN (R 4.3.3)
xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.3)
xts 0.13.2 2024-01-21 [1] CRAN (R 4.3.3)
yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.2)
yardstick * 1.3.1 2024-03-21 [1] CRAN (R 4.3.3)
zoo 1.8-12 2023-04-13 [1] CRAN (R 4.3.3)
[1] C:/Users/Willi/AppData/Local/R/win-library/4.3
[2] C:/Program Files/R/R-4.3.3/library
D ── DLL MD5 mismatch, broken installation.
──────────────────────────────────────────────────────────────────────────────